Structure and function of the TORC1 signaling network

The TORC1 signaling network mapped using functional genomics. The green node (in the center of the network) is TORC1.
The Target of Rapamycin Kinase Complex I (TORC1) is a key regulator of cell growth and metabolism in eukaryotes. In the presence of pro-growth hormones and ample nutrients, TORC1 is active and drives protein, lipid, and nucleotide synthesis by phosphorylating a wide range of proteins. In contrast, when hormone, nutrient, or energy levels fall—or cells are exposed to noxious stress—TORC1 is inhibited, causing the cell to switch from anabolic to catabolic metabolism and, eventually enter a quiescent state. Disruption of these signaling events–either upstream or downstream of TORC1–leads to numerous diseases including cancer and diabetes.
It is now well established that amino acid signals are transmitted to TORC1 via a pair of small GTPases, called Gtr1/2 in yeast and Rag A/B and C/D in humans. However, how other stress and starvation signals–such as glucose starvation, osmotic stress, oxidative stress and heat stress–are transmitted to TORC1 remains unknown. To address this question, we are using high-throughput genetic, genomic, and biochemical approaches to map the signaling pathways that regulate TORC1 in different conditions, and determine how these pathways cooperate to control cell growth and metabolism.
TORC1-body formation and the role of protein agglomeration in cell signaling
We recently discovered that TORC1 moves into a single body (containing around 200 TORC1 molecules), located on the edge of the vacuolar/lysosomal membrane during glucose and nitrogen starvation. Glucose/nitrogen repletion then cause the body to dissociate, so that TORC1 spreads back across the vacuolar/lysosomal membrane.
Following this discovery up, we showed that AMPK/Snf1 phosphorylates the key regulatory subunit in TORC1 (Kog1/Raptor) during glucose starvation at two novel sites (Ser 491 and 494), and that this phosphorylation event helps drive the formation of TORC1-bodies (TORC1-bodies form 20-fold slower in Kog1S491/494A and snf1 delete cells). Ser 491 and 494 sit in one of two glutamine-rich, prion-like, domains in Kog1--and these sequences also turned out to be very important for TORC1-body formation (TORC1-bodies form 100-fold slower in the strongest prion mutant; PrDm1+2). Finally, by studying strains with mutations that limit TORC1-body formation (Kog1S491/494A, PrDm1+2, and others) we showed that TORC1-bodies increase the threshold for TORC1 activation in cells that have been starved for a significant period of time (from around 0.02% glucose to around 2% glucose). Thus, TORC1-body formation creates hysteresis (memory) in the TORC1 pathway to help ensure that cells remain committed to a starvation state until they are exposed to optimum conditions.
We are now using a combination of genomic, genetic and biochemical approaches, in combination with live cell microscopy to determine: (1) Which signaling pathways cooperate with Snf1/AMPK to trigger TORC1-body assembly and disassembly. (2) What proteins are in the TORC1-body. (3) What physical events drive the assembly of the TORC1-body. (4) What structure the TORC1-body takes up. (5) What impact the TORC1-bodies have on cell signaling, cell growth, and the transition into quiescence.
This work should help us to determine how protein bodies--found in organisms ranging from yeast to human--modulate cell signaling and stress responses.
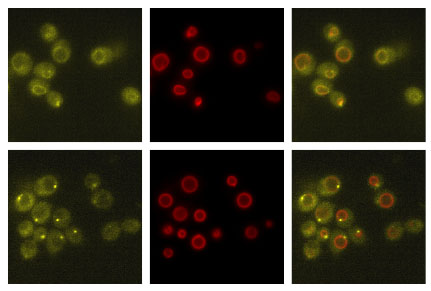
Above–Movement of Kog1-YFP (yellow) from its position distributed around the vacuolar membrane in nutrient replete conditions (upper panels) to a single body on the edge of the vacuolar membrane (lower panels). The red signal (Vph1-mCherry) marks the vacuolar membrane.
Below–Dynamics of TORC1-body formation in different stress and starvation conditions, as measured by live cell microscopy.

Cooperation between the TORC1 and other signaling pathways

Cooperation between the PIP and TORC1 Signaling pathways, mapped using genomic approaches.
While TORC1 acts as the central hub in the growth control network of eukaryotes–it certainly does not act on its own. We are therefore working to determine how TORC1 cooperates with other important signaling pathways, such as the cAMP dependent protein kinase (PKA) pathway, the AMP activated protein kinase (AMPK) pathway, inositol pyrophosphate (IP), and the protein kinase C (PKC) pathway to control cell growth, metabolism, and key developmental choices. To do this we measure gene expression in strains carrying mutations in key genes in different conditions (using RNA seq) to construct detailed, global, network models. This work will not only shed light on the structure and function of the cell growth control network in eukaryotes (and help us understand how it can go wrong), but will help us see how cells use complex signaling networks to integrate information and make decisions.
